from IPython.display import Image
Image('../../Python_probability_statistics_machine_learning_2E.png',width=200)
Logistic regression is one example of a wider class of Generalized Linear Models (GLMs). These GLMs have the following three key features
A target $Y$ variable distributed according to one of the exponential family of distributions (e.g., Normal, binomial, Poisson)
An equation that links the expected value of $Y$ with a linear combination of the observed variables (i.e., $\left\{ x_1,x_2,\ldots,x_n \right\}$).
A smooth invertible link function $g(x)$ such that $g(\mathbb{E}(Y)) = \sum_k \beta_k x_k$
Exponential Family¶
Here is the one-parameter exponential family,
The natural parameter is $\lambda$ and $y$ is the sufficient statistic. For example, for logistic regression, we have $\gamma(\lambda)=-\log(1+e^{\lambda})$ and $\lambda=\log{\frac{p}{1-p}}$.
An important property of this exponential family is that
To see this, we compute the following,
Using the same technique, we also have,
which explains the usefulness of this generalized notation for the exponential family.
Deviance¶
The scaled Kullback-Leibler divergence is called the deviance as defined below,
Hoeffding's Lemma.
Using our exponential family notation, we can write out the deviance as the following,
where $\mu_1:=\mathbb{E}_{\lambda_1}(y)$. For the maximum likelihood estimate $\hat{\lambda}_1$, we have $\mu_1=y$. Plugging this into the above equation gives the following,
Taking the negative exponential of both sides gives,
Because $D$ is always non-negative, the likelihood is maximized when the the deviance is zero. In particular, for the scalar case, it means that $y$ itself is the best maximum likelihood estimate for the mean. Also, $f(y;\hat{\lambda}_1)$ is called the saturated model. We write Hoeffding's Lemma as the following,
to emphasize that $f(y;y)$ is the likelihood function when the mean is replaced by the sample itself and $f(y;\mu)$ is the likelihood function when the mean is replaced by $\mu$.
Vectorizing Equation (2) using mutual independence gives the following,
The idea now is to minimize the deviance by deriving,
This means the the MLE $\hat{\boldsymbol{\beta}}$ is the best $p\times 1$ vector $\boldsymbol{\beta}$ that minimizes the total deviance where $g$ is the link function and $\mathbf{M}$ is the $p\times n$ structure matrix. This is the key step with GLM estimation because it reduces the number of parameters from $n$ to $p$. The structure matrix is where the associated $x_i$ data enters into the problem. Thus, GLM maximum likelihood fitting minimizes the total deviance like plain linear regression minimizes the sum of squares.
With the following,
with $2\times n$ dimensional $\mathbf{M}$. The corresponding joint density function is the following,
where
and
where now the sufficient statistic is $\boldsymbol{\xi}$ and the parameter vector is $\boldsymbol{\beta}$, which fits into our exponential family format, and $\mathbf{m}_i$ is the $i^{th}$ column of $\mathbf{M}$.
Given this joint density, we can compute the log likelihood as the following,
To maximize this likelihood, we take the derivative of this with respect to $\boldsymbol{\beta}$ to obtain the following,
since $\gamma'(\mathbf{m}_i^T \boldsymbol{\beta}) = \mathbf{m}_i^T \mu_i(\boldsymbol{\beta})$ and (c.f. Equation (1)), $\gamma' = \mu_{\lambda}$. Setting this derivative equal to zero gives the conditions for the maximum likelihood solution,
where $\boldsymbol{\mu}$ is the element-wise inverse of the link function. This leads us to exactly the same place we started: trying to regress $\mathbf{y}$ against $\boldsymbol{\mu}(\mathbf{M}^T\boldsymbol{\beta})$.
Example¶
The structure matrix $\mathbf{M}$ is where the $x_i$ data associated with the corresponding $y_i$ enters the problem. If we choose
where $\mathbf{1}$ is an $n$-length vector and
with $\mu(x) = 1/(1+e^{-x})$, we have the original logistic regression problem.
Generally, $\boldsymbol{\mu}(\boldsymbol{\beta})$ is a nonlinear function and thus we regress against our transformed variable $\mathbf{z}$
This fits the format of the Gauss Markov (see ch:stats:sec:gauss) problem and has the
following solution,
where
where $g$ is the link function and $\mathbf{v}$ is the variance function on the designated distribution of the $y_i$. Thus, $\hat{\boldsymbol{\beta}}$ has the following covariance matrix,
These results allow inferences about the estimated parameters $\hat{\boldsymbol{\beta}}$. We can easily write Equation (4) as an iteration as follow,
%matplotlib inline
from scipy.stats.distributions import poisson
import statsmodels.api as sm
from matplotlib.pylab import subplots
import numpy as np
def gen_data(n,ns=10):
param = n**2
return poisson(param).rvs(ns)
fig,ax=subplots()
xi = []
yi = []
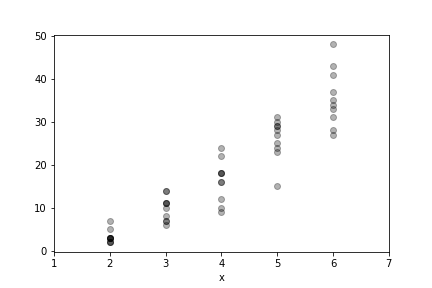
for i in [2,3,4,5,6]:
xi.append([i]*10)
yi.append(gen_data(i))
_=ax.plot(xi[-1],yi[-1],'ko',alpha=.3)
_=ax.axis(xmin=1,xmax=7)
xi = np.array(xi)
yi = np.array(yi)
x = xi.flatten()
y = yi.flatten()
_=ax.set_xlabel('x');
fig.savefig('fig-machine_learning/glm_003.png')
Some data for Poisson example.
We can use our iterative matrix-based approach. The following code initializes the iteration.
M = np.c_[x*0+1,x].T
gi = np.exp # inverse g link function
bk = np.array([.9,0.5]) # initial point
muk = gi(M.T @ bk).flatten()
Rz = np.diag(1/muk)
zk = M.T @ bk + Rz @ (y-muk)
and this next block establishes the main iteration
while abs(sum(M @ (y-muk))) > .01: # orthogonality condition as threshold
Rzi = np.linalg.inv(Rz)
bk = (np.linalg.inv(M @ Rzi @ M.T)) @ M @ Rzi @ zk
muk = gi(M.T @ bk).flatten()
Rz =np.diag(1/muk)
zk = M.T @ bk + Rz @ (y-muk)
with corresponding final $\boldsymbol{\beta}$ computed as the following,
print(bk)
with corresponding estimated $\mathbb{V}(\hat{\boldsymbol{\beta}})$ as
print(np.linalg.inv(M @ Rzi @ M.T))
The orthogonality condition Equation (3) is the following,
print(M @ (y-muk))
For comparison, the statsmodels module provides the Poisson GLM object.
Note that the reported standard error is the square root of the
diagonal elements of $\mathbb{V}(\hat{\boldsymbol{\beta}})$. A plot of
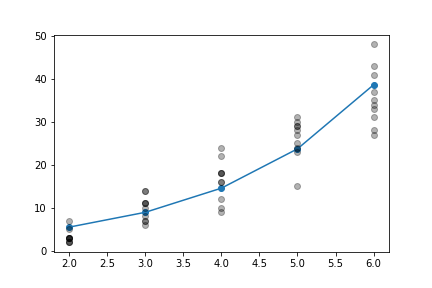
the data and the fitted model is shown below in Figure (fig:glm_004).
pm=sm.GLM(y, sm.tools.add_constant(x),
family=sm.families.Poisson())
pm_results=pm.fit()
pm_results.summary()
b0,b1=pm_results.params
fig,ax=subplots()
_=ax.plot(xi[:,0],np.exp(xi[:,0]*b1+b0),'-o');
_=ax.plot(xi,yi,'ok',alpha=.3);
fig.savefig('fig-machine_learning/glm_004.png')
_=ax.set_xlabel('x');
Fitted using the Poisson GLM.